RNA-based technologies are integral to research
Basic and clinical research have been greatly influenced by RNA-based technologies. With the discovery of gene silencing through RNA interference (RNAi), scientists could directly test gene function, using small sequences of RNA termed microRNA (miRNA) and small interfering RNA (siRNA).1 Later on, “[The] discovery [of CRISPR] has spurred an entire biotech revolution of its own”, as Nobel Prize Winner,2 Dr. Jennifer Doudna, recently reminisced in an essay she penned for The Atlantic3.
Doudna’s team, collaborators, and other scientists have harnessed the power of CRISPR to provide a likely cure for sickle-cell anemia. The first patients were treated4 during clinical trials just two years ago and remain symptom-free to date. CRISPR has also been utilized in CAR-T3,5 cell therapy to treat cancer and rare diseases in humans.3,6 In addition to therapeutic benefits, CRISPR-based technologies have been successfully applied to the veterinary and agricultural industries.3
Beyond gene editing and silencing for therapeutic use, RNA-based technologies are routinely employed in target validation for small molecule drugs and monoclonal antibody therapeutics, and to validate monoclonal and polyclonal antibodies for use across a variety of assays (Figure 1). Included in this list are anti-drug antibody assays,7,8 which are critical in interrogating the immune response to a drug. The five pillars for antibody validation include orthogonal strategies reliant on siRNA technology to knock down the target protein and confirm antibody specificity.9 Upstream, siRNA has also been used to develop small-molecule compound screens for lead discovery.10
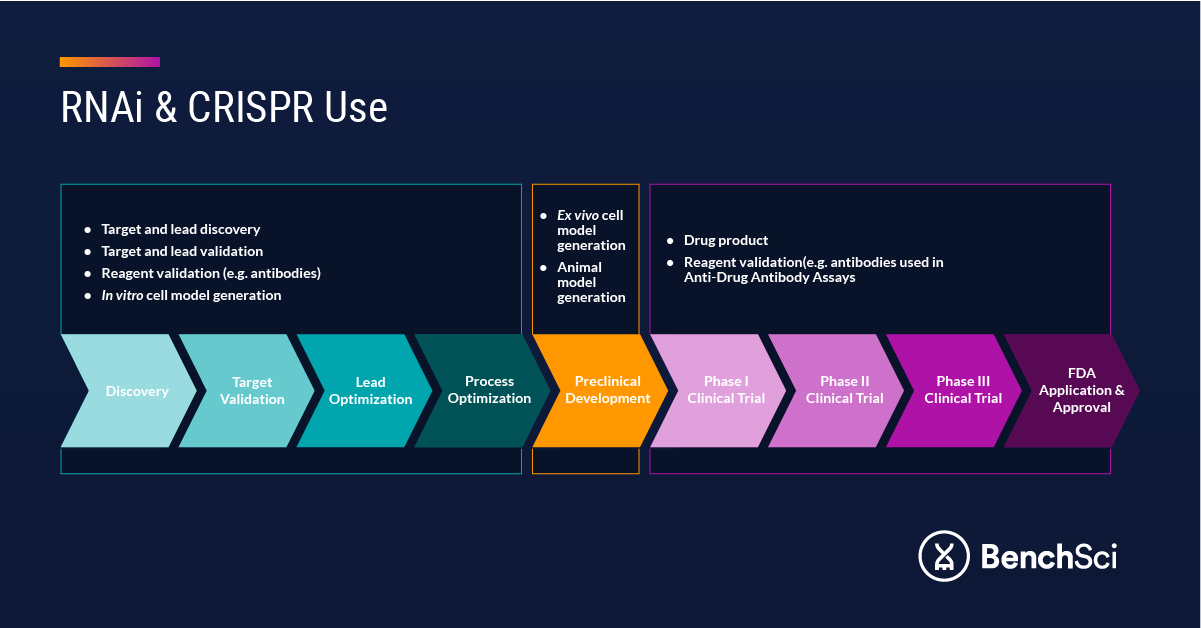 Figure 1. RNA Uses in Drug Discovery and Development.
Figure 1. RNA Uses in Drug Discovery and Development.
Off-target binding of RNA-based tools challenges research progress
Both RNAi and CRISPR use a strand of RNA to bind to a target sequence. 1 However, if there are other genetic sequences in target species that are similar to the target sequence, the reagents may bind in undesired locations. Off-target binding often impacts the expression of other genes, and subsequently leads to unanticipated or detrimental experimental results.
For both RNAi and CRISPR work, translating in vitro and ex vivo results into in vivo models may confer additional problems, as the target sequence may be similar to other species’ genetic sequences, leading to further confounding data, which can affect research at both the basic and clinical level.
To date, many studies have highlighted that off-target interactions observed with CRISPR-Cas9 are dependent on the guide RNA (gRNA) sequence and experimental conditions;11 gRNA binding activity has also been shown to be quite variable.12 In two studies published in 2014 and 2016 in Nature Biotechnology, 11,12 Doench and colleagues proposed a metric to score the efficiency (on-target activity) and cross-reactivity (off-target binding) of gRNAs.
To address some of these challenges, a new feature has been added to BenchSci’s platform to provide scientists with more comprehensive data about off-target binding. Along with vendor-provided information, you can now see how the expected target sequence of an RNAi or gRNA reagent compares with the transcriptome or genome of the target species. In addition, on-target and off-target binding scores for gRNA products are provided using the methods established by Doench et al. to predict the success and potential off-target capacity of CRISPR-Cas9 tools.
Off-target binding data sets scientists up for success
“Designing experiments with RNAi and CRISPR poses some unique challenges. You may need to use a variety of bioinformatics tools and calculations to determine if a product may inadvertently target other genes across the genome due to similarities in the sequence. This is a critical consideration because these off-target effects could greatly impact your results,” says Stephanie Prezioso, Ph.D., Group Product Manager, Reagent Selection. “That’s why we added off-target information for RNAi and gRNA reagents in-app. BenchSci can surface and flag those details at the initial stages of product selection so that scientists can begin their experiments with more confidence and have greater success down the line.”
Identifying the off-target binding potential of RNA-based reagents is important for anticipating potential experiment pitfalls. Having this data all in one place and easily visible while searching for reagents helps scientists plan experiments and research projects more effectively.
Furthermore, off-binding data on BenchSci’s platform is displayed consistently across all vendors, so scientists can easily compare products from different sources without having to consult multiple websites or bioinformatics tools prior to making a purchasing decision.
With this update, we hope scientists can feel more confident about choosing the best reagents for their experiments while pushing the boundaries of their research. Visit the Knowledge Center for more information on these and other features of the BenchSci platform.
References
- Sasso, J. M., et al. 2022. The Progress and Promise of RNA Medicine – An Arsenal of Targeted Treatments. J Med Chem 65(10): 6975-7015 https://doi.org/10.1021/acs.jmedchem.2c00024
- The Nobel Prize Organization. 2020. Press release: The Nobel Prize in Chemistry 2020. October 7, 2020. https://www.nobelprize.org/prizes/chemistry/2020/press-release/
- Doudna, J. A. Starting a Revolution isn’t enough. The Atlantic. September Issue. 2022. September 12, 2022. https://www.theatlantic.com/science/archive/2022/09/crispr-cas9-gene-editing-biotechnology/671382/
- Stein, R. First sickle cell patient treated with CRISPR gene-editing still thriving. Health News from NPR. December 31, 2021. https://www.npr.org/sections/health-shots/2021/12/31/1067400512/first-sickle-cell-patient-treated-with-crispr-gene-editing-still-thriving
- LaHucik, K. Intellia’s CRISPR-engineered cell therapy secures FDA orphan drug status after AML trial launch. Fierce Biotech. March 9, 2022. https://www.fiercebiotech.com/biotech/intellias-1st-ex-vivo-cell-therapy-aml-secures-fda-orphan-drug-status-week-after-trial
- Businesswire. Cure Rare Disease Receives FDA Approval to Administer First-in-Human CRISPR Therapeutic. August 10, 2022. https://www.businesswire.com/news/home/20220810005035/en/Cure-Rare-Disease-Receives-FDA-Approval-to-Administer-First-in-Human-CRISPR-Therapeutic
- Wang, J. Development of Multiplex Sensitivity Anti-Drug Antibody Assays for CRISPR/Cas9 Gene Therapies. Editas Medicine. Presentation. September 27, 2017. https://www.editasmedicine.com/wp-content/uploads/2019/10/immunogenicity_final_1508786835.pdf
- Simhadri, V.L., et al. 2018. Mol Ther Methods Clin Dev 10: 105-112, doi: 10.1016/j.omtm.2018.06.006
- Uhlen, M., et al. 2016. A proposal for validation of antibodies. Nature Methods 13: 823-827. https://doi.org/10.1038/nmeth.3995
- Ding, M., et al. 2020. Combined siRNA and Small-Molecule Phenotypic Screening Identifies Targets Regulating Rhinovirus Replication in Primary Human Bronchial Epithelial Cells. SLAS 25(6) https://doi.org/10.1177/247255522090972
- Doench, J. G., et al. 2014. Rational Design of Highly Active sgRNAs for CRISPR-Cas9-Mediated Gene Activation. Nat Biotechnol 32(12): 1262-1267 doi: 10.1038/nbt.3026
- Doench, J. G., et al. 2016. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol 34: 184-191 https://doi.org/10.1038/nbt.3437